Difference between revisions of "Biomedical Forensics"
Jbringardner (talk | contribs) |
|||
| (206 intermediate revisions by 13 users not shown) | |||
| Line 1: | Line 1: | ||
= | = Objective = | ||
The | The objective of this experiment is to use the biomedical forensic techniques of extracting DNA, fingerprinting, identifying foreign substances, and blood typing to investigate a crime scene. | ||
= Overview = | = Overview = | ||
Biomedical forensic methods are used in criminal or civil cases. They are also used within the medical field to diagnose and treat diseases and injuries. Within criminal law, these techniques are used to identify suspects in criminal cases and to exclude individuals as suspects. DNA testing, in particular, is increasingly used to prove the innocence of people who have been wrongfully convicted of a crime. | |||
In this lab, DNA extraction will be introduced using basic biochemical techniques for isolating, purifying, and digesting DNA molecules. A toxicology report will be completed by observing chemical reactions using simulated reagents. A simulation kit will be used for the blood typing tests. | |||
== DNA Extraction == | |||
<b>Deoxyribonucleic acid (DNA)</b> is found in almost all living organisms. In this lab, DNA will be extracted from plant cells in an introductory exercise to this lab. | |||
Plant cells are surrounded by structures called a cell wall and plasma membrane that protect the cell. Inside the cell sits various units called organelles; DNA is located in an organelle called the nucleus. | |||
The <b>DNA extraction</b> process can be divided into three major steps: breaking open the cell wall, destroying membranes within the cell, and precipitating the DNA out of the solution. | |||
The first step in DNA extraction is to <b>break the cell wall (cell lysis)</b> to expose the DNA within the cell. Cell walls are rigid structures made of cellulose and thus require mechanical methods to break apart. In this experiment, DNA will be extracted from a strawberry that will be mashed manually. | |||
The second step in DNA extraction is to <b>destroy the plasma membrane within the cell</b>. The cell's plasma membrane is made of phospholipid bilayers that are made of fat. To disrupt them, the mesh of fat molecules is broken up with a surfactant (soap). The structure of soap is similar to the structure of fat and grease and lowers the surface tension between two fluids when acting as a surfactant. | |||
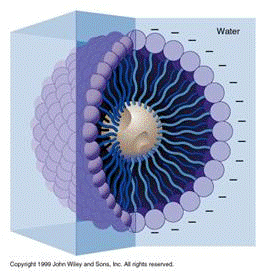
The | A soap molecule has two parts: a head and a tail. The head is polar and is attracted to water (hydrophilic), while the tail is nonpolar and attracted to oil and fat (hydrophobic). When soap molecules are in water, they group themselves into micelles that have a roughly spherical structure in which all the polar heads point outwards (toward water) and all the nonpolar tails point inwards at the center of the sphere (away from the water) (Figure 5). | ||
[[Image:DNA4.gif|thumb|500px|center|Figure 5: Soap Micelle]] | |||
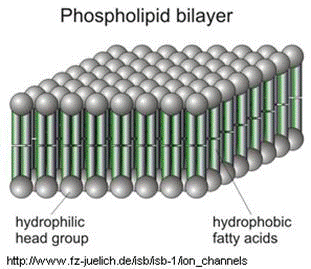
The soap molecules effectively trap the fat molecules inside the micelles and dissolve the cell membranes. The soap molecules orient themselves so that their head associates with the tail of the phospholipid bilayer. In this way, the soap breaks up the bilayer molecule by molecule (Figure 6). | |||
[[Image:DNA14.gif|thumb|500px|center|Figure 6: Phospholipid Bilayer]] | |||
The last step in DNA extraction is <b>precipitating the DNA</b>. When the plasma membrane is successfully disrupted, the DNA is released from the cells into the solution along with protein molecules and other cellular miscellany. With the cell's contents mixed into a solution, the DNA can be separated from the rest of the contents. This process is called precipitation. Salt is used because it disrupts the structure of the proteins and carbohydrates found in the solution. Also, the salt provides a favorable environment to extract the DNA by contributing positively-charged sodium ions that neutralize the negative charge of DNA. | |||
After adding salt and soap, the extracted DNA cannot be seen as it is too small to distinguish from the rest of the solution. To make the DNA visible, alcohol is added since it cannot dissolve the DNA. A translucent white substance will begin to form at the top: this is the DNA (Figure 7). Once the precipitate is thick enough, it can be spooled out. This simple procedure is a rough extraction process that needs further steps before the DNA can be successfully run on a gel for analysis. This analysis will not be performed in this lab. | |||
[[Image: | [[Image:DNA5.gif|frame|500px|center|Figure 7: Extracted DNA]] | ||
== | == Fingerprints == | ||
<b>Fingerprints</b> are unique patterns consisting of friction ridges (raised) and furrows (recessed) that appear on the pads of the fingers and thumbs and are often used for identification purposes in forensics. Prints from palms, toes, and feet are also unique, but these are used less often for identification so this guide focuses on prints from the fingers and thumbs. | |||
The fingerprint pattern, such as the print left when an inked finger is pressed onto paper, is made from the friction ridges on that particular finger. Friction ridge patterns are grouped into three distinct types – loops, whorls, and arches – each with unique variations, depending on the shape and relationship of the ridges. | |||
<b>Loops</b> are prints that recurve back on themselves to form a loop shape (Figure 8). Divided into radial loops (pointing toward the radius bone, or thumb) and ulnar loops (pointing toward the ulna bone, or pinky), loops account for approximately 60% of pattern types. | |||
[[Image:Loops.png|thumb|500px|center|Figure 8: Loop Fingerprint]] | |||
<b>Whorls</b> form circular or spiral patterns, like tiny whirlpools (Figure 9). There are four groups of whorls: plain (concentric circles), central pocket loop (a loop with a whorl at the end), double loop (two loops that create an S-like pattern) and accidental loop (irregularly shaped). Whorls make up about 35% of pattern types. | |||
[[Image: | [[Image:Whorls.png|thumb|500px|center|Figure 9: Whorl Fingerprint]] | ||
<b>Arches</b> create a wave-like pattern and include plain arches and tented arches (Figure 10). Tented arches rise to a sharper point than plain arches. Arches make up about 5% of all pattern types. | |||
[[Image:arches.png|thumb|500px|center|Figure 10: Arch Fingerprint]] | |||
[[Image: | |||
== Blood Typing == | |||
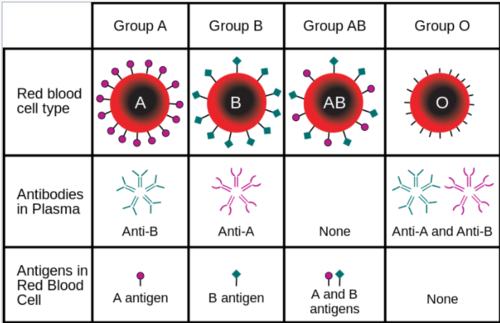
Even though all red blood cells are made of similar elements, not all blood cells are alike. There are eight different types of blood, each based on the presence or absence of three types of <b>antigens</b> (A, B, and Rh) (Figure 11). Generally, antigens induce an immune response to foreign substances in the body. The <b>A and B antigens</b> determine the type of blood while the <b>Rh antigen</b> determines if the blood is positive or negative with the Rhesus factor, which is associated with hemolytic diseases and incompatible blood transfusions. | |||
[[Image: | *The ABO Blood Group System | ||
**Group A has only the A antigen on red cells | |||
**Group B has only the B antigen on red cells | |||
**Group AB has both A and B antigens on red cells | |||
**Group O has neither A nor B antigens on red cells | |||
Positive (+) blood types have the Rh factor and negative (-) blood types do not have the Rh factor. | |||
[[Image:Lab_biomed_1.PNG|thumb|500px|frame|center|Figure 11: Different Blood Types]] | |||
= Materials and Equipment = | = Materials and Equipment = | ||
< | *Strawberry | ||
*Non-iodized table salt (NaCl) | |||
*Hand soap (clear, unscented) | |||
*95% isopropyl alcohol (0 °C) | |||
*Distilled water | |||
*Strainer | |||
*Plastic cups | |||
*Ziploc bag | |||
*Iron magnetic wand | |||
*Iron powder | |||
*Blood typing kit | |||
*Pipettes | |||
*Beaker | |||
*Test tubes | |||
*Microcentrifuge tubes | |||
<!--*Plexiglass pieces--> | |||
*Baby wipes | |||
*Meter stick | |||
= Procedure = | = Procedure = | ||
Watch the following video: [https://drive.google.com/file/d/18f1tHzFJg5bnU5ky9-HWcFqvm9FePp_K/view?usp=sharing Biomedical Forensics Part 1] | |||
== Forensic Academy == | |||
Basic training in three forensic techniques will be completed before the crime scene investigation is conducted. | |||
=== 1. DNA Extraction === | |||
# Place 200 ml of distilled water in a beaker. | |||
# Add 2 tsp (10 ml) of dish soap to the water. | |||
# Stir in ¼ tsp of salt and mix until the salt dissolves. This is the extraction mixture. | |||
# Place one strawberry into a Ziploc bag. | |||
# Pour the extraction mixture into the bag with the strawberry. | |||
# Remove as much air from the bag as possible and seal it closed. | |||
# Mash the strawberry inside the bag manually. Crush any large pieces. | |||
# | # Pour the resulting strawberry pulp and extraction mixture through a strainer and into a beaker. | ||
# | # Use a plastic spoon to press the mashed bits of strawberry against the strainer forcing as much of the mixture as possible into the container. | ||
# | # Pour the extraction mixture into a test tube. This will help isolate the DNA on the surface of the mixture. | ||
# | # Add 1 tsp (5 ml) of the chilled isopropyl alcohol to the solution (request the isopropyl alcohol from a TA) and hold the mixture at eye level. Look for the separation of DNA that shows up as a white layer on the surface of the mixture. | ||
# | # If the separation of DNA cannot immediately be seen, set the small container aside for a few minutes and examine it again later. | ||
# | |||
# | === 2. Fingerprinting === | ||
# Add | # Obtain a clean piece of Plexiglass and thoroughly clean it with a baby wipe. | ||
# | # Apply a thumb onto the glass and put pressure onto the glass while holding the glass by its edges. Keep applying pressure for a few seconds. | ||
# Hold the glass against the light to view the imprint to determine if it is visible. If not, clean the glass and repeat the process. | |||
# | # Take the iron filament and spread a generous amount on the region of the imprint. | ||
# | # Take the magnetic wand and slowly pull the top across the iron filament to extract the excess iron filament from the Plexiglass piece. | ||
# | # Place the excess iron filament back into the container and seal the container. | ||
# | # Closely observe the remaining fingerprint and identify the fingerprint type. | ||
# | # Take a picture of the fingerprint on a white background. | ||
# | |||
# | === 3. Toxicology Report === | ||
# | |||
# | # Obtain samples of the suspect drug from a TA. | ||
#: | # Separate the drug sample into five microcentrifuge tubes and separately label the microcentrifuge tubes A, B, C, D, or E. | ||
# | # Obtain the five drug reagents from a TA. The reagents test for the following drugs. | ||
# | #: <br> | ||
# | #: Drug A: Cannabis | ||
#: Drug B: Heroin | |||
#: Drug C: MDMA (Ecstasy) | |||
#: Drug D: Cocaine | |||
#: Drug E: Lysergic Acid Diethylamide (LSD) | |||
# | # Take the A-labelled microcentrifuge tube with the drug sample and place five droplets of the cannabis reagent. | ||
# Observe the test tube. If a reaction is observed from adding the reagent, the tube contains that drug. The microcentrifuge tube might need a quick shake to catalyze the reaction. | |||
# | # Repeat Steps 4 and 5 for each of the drug reagents. | ||
# | |||
# | |||
# | <b>Congratulations, the Forensic Academy is completed and the graduates are certified to participate in the crime scene investigation!</b> | ||
# | == Crime Scene Investigation == | ||
# | |||
# | It was a warm and stormy night on September 5, 1975. The EG1004 office was quiet as one TA was pulling a late night shift to prepare the classroom for the next morning’s lab. There was a loud crash in the background. The room went dark. The next morning, there was police tape barring entry to the entire room. Someone had murdered Ruhit Roy. At the crime scene, police discovered a pool of blood, along with the suspected murder weapon – a shard of glass – near the body. When the investigation yielded no results, Ruhit’s murder was declared a cold case. Watch the following video: [https://drive.google.com/file/d/15w4Wzz0CZ6b40Don5Qm-U--HXUYZ35i5/view?usp=sharing Biomedical Forensics Part 2] | ||
# | |||
# | After many years, Ruhit’s case is finally being reopened. Watch the following video: [https://drive.google.com/file/d/1QHC4Wt8yRMiDP3if-xIbV7D5mQergxF4/view?usp=sharing Biomedical Forensics Part 3] | ||
# | <b>Download the [https://docs.google.com/spreadsheets/d/10Mmt_5Xxx_TjEqNTz_aRtXNUEmbcAcXj/edit?usp=sharing&ouid=100215762625371235485&rtpof=true&sd=true Suspect Information Sheet] to get started on the investigation.</b> | ||
# | |||
# | === 1. Biological Dimensional Analysis === | ||
# | # Obtain a yard stick and measure the length of a foot with shoes on. | ||
# | # Measure the length of a hand from the tip of the middle finger to the bottom of the palm. | ||
# | # Repeat Steps 1 and 2 for the additional members in the group. | ||
# Calculate and compare the ratios of the hand length to foot length and determine the average foot to hand length ratio based on the data obtained. | |||
# Using the average foot length to hand length ratio, approximate the hand length of the murderer using the 11 in footprint found at the crime scene. Record the estimated measurements of the killer (Figure 12). | |||
[[Image:Foot.png|thumb|600px|center|Figure 12: An 11" Footprint Left at the Crime Scene]] | |||
=== 2. Toxicology Report === | |||
# To collect important information on the murder, the victim’s blood will be tested to determine the drug that killed him. | |||
# Put a small sample of Roy’s blood into five microcentrifuge tubes. Use the drug reagents to determine which drug was injected into his bloodstream by the killer. | |||
# Record the drug that was used on the victim. | |||
<font color="red"><b>'''Note: Please use caution when using the microcentrifuge tubes, as they can explode.'''</b></font> | |||
Watch the following video: [https://drive.google.com/file/d/1yCpCj2oNPDNIy-90OfRXMC4ZazPpiMRk/view?usp=sharing Biomedical Forensics Part 4] | |||
=== 3. Blood Typing === | |||
# Obtain the blood sample from the crime scene and from all 10 suspects and the victim along with their respective A, B, and Rh serum capsules. | |||
# Using a pipette, administer three drops of Suspect 1’s blood into all three ovals of the test tray. | |||
# Using a pipette, administer a one drop of the A serum into the oval labeled A, administer one drop of the B serum into the oval labeled B, and administer one drop of the Rh serum into the oval labeled Rh. Wash the pipette out with water after administering each serum and between each suspect to prevent cross-contamination. | |||
# If the blood sample exhibits a reaction to the indicator (bubbles or becomes cloudy), then the blood type matches that indicator type. If the liquid appears to be more diluted (more watery or thin), then the blood type does not match that indicator type. For example, if a precipitate forms for A and Rh for a blood sample, then it is A+. If a precipitate forms for A and B, but not Rh, that blood type is AB- If the blood samples do not react to any serum, then that blood type is O-. | |||
# If a precipitate forms for A and B, but not Rh, that person’s blood type is AB-. | |||
# Repeat Steps 2-5 for each suspect. Do <b>not</b> mix up the serums as that will determine the wrong blood type. | |||
# The blood found at the scene of the crime was A-. Test Roy’s blood to make sure that the blood at the scene is the killer’s blood and not Roy’s blood. | |||
# Add the blood types of each person to the Suspect Information Sheet to determine if any suspect’s blood type matches the blood type found at the crime scene. | |||
=== 4. Fingerprint Test === | |||
# Carefully obtain the murder weapon (the shard of glass). | |||
# Carefully apply the iron filament at the rim of the glass shard, trying not to waste filament. | |||
# Using the magnetic wand, carefully remove the excess filament. | |||
# Fingerprint residue should be left behind after removing the excess filament; if not, repeat Steps 2 and 3. | |||
# Observe the fingerprint, noting its features and type. Determine which person it corresponds to in the Suspect Information Sheet. | |||
Using all the information gathered in the analyses, use the data provided in the Suspect Information Sheet to determine who the killer is. Tell a TA who the alleged killer is. | |||
= Assignment = | = Assignment = | ||
== | == There is no lab report for Lab 11. == | ||
* | <!--{{Labs:Lab Report}} | ||
* Justify the use of salt, soap, and alcohol in the extraction procedure | |||
* | * Discuss the importance of biomedical forensics | ||
* | * Explain the three steps of DNA extraction | ||
* Describe the most common types of fingerprints | |||
* Clearly describe the procedural steps | * Explain what causes different blood types | ||
* Describe the steps carried out with the TA | * Justify the use of salt, soap, and alcohol in the extraction procedure | ||
* | * Discuss how knowing the victim's blood type helped find the killer | ||
* Include | * Discuss the important properties of DNA directly having an impact on the extraction procedure | ||
* Clearly describe the procedural steps that were carried out in lab | |||
* Describe the steps carried out with the TA | |||
* Include pictures of the extracted DNA, fingerprint on the murder weapon, and drug present in the victim’s blood | |||
* Include the hand size and blood type of the killer | |||
* How would procedure change if meat was used instead of fruit? | * How would procedure change if meat was used instead of fruit? | ||
* Discuss improvements that could be made to the experiment | |||
* Discuss what part of the lab each individual member completed for the group and how it was important to the overall experiment. | |||
{{Lab | {{Labs:Lab Notes}}--> | ||
== Team PowerPoint Presentation == | == Team PowerPoint Presentation == | ||
Author a presentation using (nearly) all images. The following areas are where text is allowed: | |||
* Rely heavily on graphics and pictures | * Title slide | ||
* Make sure the | * Overview slide | ||
* Discuss the real-life application of DNA | * Materials slide | ||
* Demonstrate clear understanding of each procedural step carried out and why it worked | * Captions for figures, tables, and equations | ||
* Suspect list/table | |||
* Data tables | |||
* References to suspect names | |||
'''Text is allowed for slide titles''' | |||
{{Labs:Team Presentation}} | |||
* Rely heavily on graphics and pictures | |||
* Make sure the experimental work is described simply and thoroughly | |||
* Discuss the real-life application of DNA extraction | |||
* Demonstrate clear understanding of each procedural step carried out and why it worked | |||
= References = | = References = | ||
<p>[http://www.vernier.com http://www.vernier.com]</p> | <p>[http://www.vernier.com http://www.vernier.com]</p> | ||
<p>[http://www.invitrogen.com http://www.invitrogen.com]</p> | <p>[http://www.invitrogen.com http://www.invitrogen.com]</p> | ||
<p>[http://en.wikipedia.org http://en.wikipedia.org]</p> | <p>[http://en.wikipedia.org http://en.wikipedia.org]</p> | ||
{{Laboratory Experiments}} | {{Laboratory Experiments}} | ||
Latest revision as of 19:39, 18 April 2024
Objective
The objective of this experiment is to use the biomedical forensic techniques of extracting DNA, fingerprinting, identifying foreign substances, and blood typing to investigate a crime scene.
Overview
Biomedical forensic methods are used in criminal or civil cases. They are also used within the medical field to diagnose and treat diseases and injuries. Within criminal law, these techniques are used to identify suspects in criminal cases and to exclude individuals as suspects. DNA testing, in particular, is increasingly used to prove the innocence of people who have been wrongfully convicted of a crime.
In this lab, DNA extraction will be introduced using basic biochemical techniques for isolating, purifying, and digesting DNA molecules. A toxicology report will be completed by observing chemical reactions using simulated reagents. A simulation kit will be used for the blood typing tests.
DNA Extraction
Deoxyribonucleic acid (DNA) is found in almost all living organisms. In this lab, DNA will be extracted from plant cells in an introductory exercise to this lab.
Plant cells are surrounded by structures called a cell wall and plasma membrane that protect the cell. Inside the cell sits various units called organelles; DNA is located in an organelle called the nucleus.
The DNA extraction process can be divided into three major steps: breaking open the cell wall, destroying membranes within the cell, and precipitating the DNA out of the solution.
The first step in DNA extraction is to break the cell wall (cell lysis) to expose the DNA within the cell. Cell walls are rigid structures made of cellulose and thus require mechanical methods to break apart. In this experiment, DNA will be extracted from a strawberry that will be mashed manually.
The second step in DNA extraction is to destroy the plasma membrane within the cell. The cell's plasma membrane is made of phospholipid bilayers that are made of fat. To disrupt them, the mesh of fat molecules is broken up with a surfactant (soap). The structure of soap is similar to the structure of fat and grease and lowers the surface tension between two fluids when acting as a surfactant.
A soap molecule has two parts: a head and a tail. The head is polar and is attracted to water (hydrophilic), while the tail is nonpolar and attracted to oil and fat (hydrophobic). When soap molecules are in water, they group themselves into micelles that have a roughly spherical structure in which all the polar heads point outwards (toward water) and all the nonpolar tails point inwards at the center of the sphere (away from the water) (Figure 5).
The soap molecules effectively trap the fat molecules inside the micelles and dissolve the cell membranes. The soap molecules orient themselves so that their head associates with the tail of the phospholipid bilayer. In this way, the soap breaks up the bilayer molecule by molecule (Figure 6).
The last step in DNA extraction is precipitating the DNA. When the plasma membrane is successfully disrupted, the DNA is released from the cells into the solution along with protein molecules and other cellular miscellany. With the cell's contents mixed into a solution, the DNA can be separated from the rest of the contents. This process is called precipitation. Salt is used because it disrupts the structure of the proteins and carbohydrates found in the solution. Also, the salt provides a favorable environment to extract the DNA by contributing positively-charged sodium ions that neutralize the negative charge of DNA.
After adding salt and soap, the extracted DNA cannot be seen as it is too small to distinguish from the rest of the solution. To make the DNA visible, alcohol is added since it cannot dissolve the DNA. A translucent white substance will begin to form at the top: this is the DNA (Figure 7). Once the precipitate is thick enough, it can be spooled out. This simple procedure is a rough extraction process that needs further steps before the DNA can be successfully run on a gel for analysis. This analysis will not be performed in this lab.
Fingerprints
Fingerprints are unique patterns consisting of friction ridges (raised) and furrows (recessed) that appear on the pads of the fingers and thumbs and are often used for identification purposes in forensics. Prints from palms, toes, and feet are also unique, but these are used less often for identification so this guide focuses on prints from the fingers and thumbs.
The fingerprint pattern, such as the print left when an inked finger is pressed onto paper, is made from the friction ridges on that particular finger. Friction ridge patterns are grouped into three distinct types – loops, whorls, and arches – each with unique variations, depending on the shape and relationship of the ridges.
Loops are prints that recurve back on themselves to form a loop shape (Figure 8). Divided into radial loops (pointing toward the radius bone, or thumb) and ulnar loops (pointing toward the ulna bone, or pinky), loops account for approximately 60% of pattern types.
Whorls form circular or spiral patterns, like tiny whirlpools (Figure 9). There are four groups of whorls: plain (concentric circles), central pocket loop (a loop with a whorl at the end), double loop (two loops that create an S-like pattern) and accidental loop (irregularly shaped). Whorls make up about 35% of pattern types.
Arches create a wave-like pattern and include plain arches and tented arches (Figure 10). Tented arches rise to a sharper point than plain arches. Arches make up about 5% of all pattern types.
Blood Typing
Even though all red blood cells are made of similar elements, not all blood cells are alike. There are eight different types of blood, each based on the presence or absence of three types of antigens (A, B, and Rh) (Figure 11). Generally, antigens induce an immune response to foreign substances in the body. The A and B antigens determine the type of blood while the Rh antigen determines if the blood is positive or negative with the Rhesus factor, which is associated with hemolytic diseases and incompatible blood transfusions.
- The ABO Blood Group System
- Group A has only the A antigen on red cells
- Group B has only the B antigen on red cells
- Group AB has both A and B antigens on red cells
- Group O has neither A nor B antigens on red cells
Positive (+) blood types have the Rh factor and negative (-) blood types do not have the Rh factor.
Materials and Equipment
- Strawberry
- Non-iodized table salt (NaCl)
- Hand soap (clear, unscented)
- 95% isopropyl alcohol (0 °C)
- Distilled water
- Strainer
- Plastic cups
- Ziploc bag
- Iron magnetic wand
- Iron powder
- Blood typing kit
- Pipettes
- Beaker
- Test tubes
- Microcentrifuge tubes
- Baby wipes
- Meter stick
Procedure
Watch the following video: Biomedical Forensics Part 1
Forensic Academy
Basic training in three forensic techniques will be completed before the crime scene investigation is conducted.
1. DNA Extraction
- Place 200 ml of distilled water in a beaker.
- Add 2 tsp (10 ml) of dish soap to the water.
- Stir in ¼ tsp of salt and mix until the salt dissolves. This is the extraction mixture.
- Place one strawberry into a Ziploc bag.
- Pour the extraction mixture into the bag with the strawberry.
- Remove as much air from the bag as possible and seal it closed.
- Mash the strawberry inside the bag manually. Crush any large pieces.
- Pour the resulting strawberry pulp and extraction mixture through a strainer and into a beaker.
- Use a plastic spoon to press the mashed bits of strawberry against the strainer forcing as much of the mixture as possible into the container.
- Pour the extraction mixture into a test tube. This will help isolate the DNA on the surface of the mixture.
- Add 1 tsp (5 ml) of the chilled isopropyl alcohol to the solution (request the isopropyl alcohol from a TA) and hold the mixture at eye level. Look for the separation of DNA that shows up as a white layer on the surface of the mixture.
- If the separation of DNA cannot immediately be seen, set the small container aside for a few minutes and examine it again later.
2. Fingerprinting
- Obtain a clean piece of Plexiglass and thoroughly clean it with a baby wipe.
- Apply a thumb onto the glass and put pressure onto the glass while holding the glass by its edges. Keep applying pressure for a few seconds.
- Hold the glass against the light to view the imprint to determine if it is visible. If not, clean the glass and repeat the process.
- Take the iron filament and spread a generous amount on the region of the imprint.
- Take the magnetic wand and slowly pull the top across the iron filament to extract the excess iron filament from the Plexiglass piece.
- Place the excess iron filament back into the container and seal the container.
- Closely observe the remaining fingerprint and identify the fingerprint type.
- Take a picture of the fingerprint on a white background.
3. Toxicology Report
- Obtain samples of the suspect drug from a TA.
- Separate the drug sample into five microcentrifuge tubes and separately label the microcentrifuge tubes A, B, C, D, or E.
- Obtain the five drug reagents from a TA. The reagents test for the following drugs.
- Drug A: Cannabis
- Drug B: Heroin
- Drug C: MDMA (Ecstasy)
- Drug D: Cocaine
- Drug E: Lysergic Acid Diethylamide (LSD)
- Take the A-labelled microcentrifuge tube with the drug sample and place five droplets of the cannabis reagent.
- Observe the test tube. If a reaction is observed from adding the reagent, the tube contains that drug. The microcentrifuge tube might need a quick shake to catalyze the reaction.
- Repeat Steps 4 and 5 for each of the drug reagents.
Congratulations, the Forensic Academy is completed and the graduates are certified to participate in the crime scene investigation!
Crime Scene Investigation
It was a warm and stormy night on September 5, 1975. The EG1004 office was quiet as one TA was pulling a late night shift to prepare the classroom for the next morning’s lab. There was a loud crash in the background. The room went dark. The next morning, there was police tape barring entry to the entire room. Someone had murdered Ruhit Roy. At the crime scene, police discovered a pool of blood, along with the suspected murder weapon – a shard of glass – near the body. When the investigation yielded no results, Ruhit’s murder was declared a cold case. Watch the following video: Biomedical Forensics Part 2
After many years, Ruhit’s case is finally being reopened. Watch the following video: Biomedical Forensics Part 3
Download the Suspect Information Sheet to get started on the investigation.
1. Biological Dimensional Analysis
- Obtain a yard stick and measure the length of a foot with shoes on.
- Measure the length of a hand from the tip of the middle finger to the bottom of the palm.
- Repeat Steps 1 and 2 for the additional members in the group.
- Calculate and compare the ratios of the hand length to foot length and determine the average foot to hand length ratio based on the data obtained.
- Using the average foot length to hand length ratio, approximate the hand length of the murderer using the 11 in footprint found at the crime scene. Record the estimated measurements of the killer (Figure 12).
2. Toxicology Report
- To collect important information on the murder, the victim’s blood will be tested to determine the drug that killed him.
- Put a small sample of Roy’s blood into five microcentrifuge tubes. Use the drug reagents to determine which drug was injected into his bloodstream by the killer.
- Record the drug that was used on the victim.
Note: Please use caution when using the microcentrifuge tubes, as they can explode.
Watch the following video: Biomedical Forensics Part 4
3. Blood Typing
- Obtain the blood sample from the crime scene and from all 10 suspects and the victim along with their respective A, B, and Rh serum capsules.
- Using a pipette, administer three drops of Suspect 1’s blood into all three ovals of the test tray.
- Using a pipette, administer a one drop of the A serum into the oval labeled A, administer one drop of the B serum into the oval labeled B, and administer one drop of the Rh serum into the oval labeled Rh. Wash the pipette out with water after administering each serum and between each suspect to prevent cross-contamination.
- If the blood sample exhibits a reaction to the indicator (bubbles or becomes cloudy), then the blood type matches that indicator type. If the liquid appears to be more diluted (more watery or thin), then the blood type does not match that indicator type. For example, if a precipitate forms for A and Rh for a blood sample, then it is A+. If a precipitate forms for A and B, but not Rh, that blood type is AB- If the blood samples do not react to any serum, then that blood type is O-.
- If a precipitate forms for A and B, but not Rh, that person’s blood type is AB-.
- Repeat Steps 2-5 for each suspect. Do not mix up the serums as that will determine the wrong blood type.
- The blood found at the scene of the crime was A-. Test Roy’s blood to make sure that the blood at the scene is the killer’s blood and not Roy’s blood.
- Add the blood types of each person to the Suspect Information Sheet to determine if any suspect’s blood type matches the blood type found at the crime scene.
4. Fingerprint Test
- Carefully obtain the murder weapon (the shard of glass).
- Carefully apply the iron filament at the rim of the glass shard, trying not to waste filament.
- Using the magnetic wand, carefully remove the excess filament.
- Fingerprint residue should be left behind after removing the excess filament; if not, repeat Steps 2 and 3.
- Observe the fingerprint, noting its features and type. Determine which person it corresponds to in the Suspect Information Sheet.
Using all the information gathered in the analyses, use the data provided in the Suspect Information Sheet to determine who the killer is. Tell a TA who the alleged killer is.
Assignment
There is no lab report for Lab 11.
Team PowerPoint Presentation
Author a presentation using (nearly) all images. The following areas are where text is allowed:
- Title slide
- Overview slide
- Materials slide
- Captions for figures, tables, and equations
- Suspect list/table
- Data tables
- References to suspect names
Text is allowed for slide titles
Follow the presentation guidelines laid out in the EG1004 Lab Presentation Format in the Technical Presentations section of the manual. When preparing the presentation, consider the following points.
- Rely heavily on graphics and pictures
- Make sure the experimental work is described simply and thoroughly
- Discuss the real-life application of DNA extraction
- Demonstrate clear understanding of each procedural step carried out and why it worked
References
| ||||||||