DNA Extraction and Gel Analysis
DNA EXTRACTION AND GEL ANALYSIS
1 OBJECTIVES
The objective of this experiment is to extract DNA from a fruit and visualize the banding pattern formed when the samples are run through a gel matrix. In doing so, you will also learn where DNA is located, how to get to it and extract it. Basic biochemical techniques will be introduced to allow you to reach, isolate, purify and digest DNA molecules.
You will also learn how to perform agarose gel electrophoresis in order to separate DNA fragments obtained from the digested samples. After the experiment, you should be able to distinguish between two different fruits by looking at the respective banding pattern.
2 OVERVIEW
Cellular Biology and Location of DNA
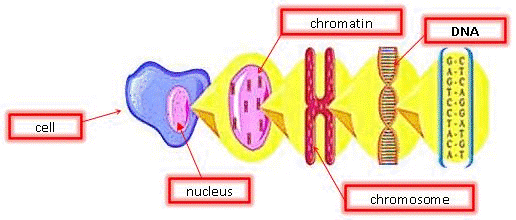
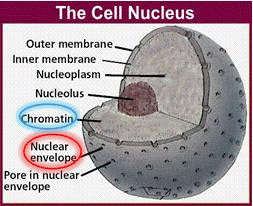
DNA is the blueprint of life. It is present in all living organisms. These organisms can be made of a single cell (bacteria) or trillions (approximately 50 trillion in the human body). There are two different types of cells: prokaryotes and eukaryotes. Prokaryotic cells do not have a nuclear membrane and thus do not have a distinct nucleus. In this experiment, we are only interested in eukaryotic cells, which make up plant and animal bodies. Eukaryotic cells contain a distinct, membrane-bound nucleus that isolates the DNA from the rest of the cell. The structure of plant cells is different from those of animal cells. We will only be dealing with plant cells in this experiment.
Plant cells are surrounded by a cell wall. It has high mechanical strength and protects the cell. Directly beneath the cell wall lies the plasma membrane which contains the cytosol. Within the cytosol along with the rest of the cellular organelles is the nucleus enclosed in the nuclear membrane. The nucleus is of particular interest for us since it houses the DNA in the form of chromatin. Chromatin is the active form of DNA in the cell when it is not preparing for cell division. It is comprised of DNA wrapped around protein particles called histones.
DNA Extraction Technique
In this experiment, one of our goals is to extract the DNA from the fruit sample. Below we will describe the scientific background behind DNA extraction.
The DNA extraction process is a fairly simple biochemical procedure that can be divided into three major steps: breaking open the cell (lysis), destroying membranes within the cell and precipitating the DNA out of the solution.
We will describe in the section below how each step relates to the physical and biochemical properties of DNA.
Cell Lysis (Breaking Open the Cell Wall and Membranes)
We have mentioned earlier that plant cells have a very rigid external structure –the cell wall – which protects it. To get to the DNA the very first step would be to break open that wall. How to do so?
The cell wall is the first barrier in our journey inside the cell towards the DNA molecule. It has a high mechanical strength – very rigid – and acts as a protector and filter. It is made of cellulose, and is the reason why wood is so hard and durable. To destroy the cell wall, we need some mechanical means in order to break apart the cellulose molecules. In our experiment, we will smash the fruit with the use of a blender.
Destroying Membranes Within the Cell
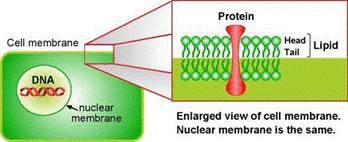
In order to know how to break something, you will most likely wonder what it is made of. For example, if you want to break through a wooden gate or a steel gate, you might consider different techniques. The same kind of critical thinking is needed here. The various membranes inside the cell are made of phospholipid bilayers. Don’t mind the scary biochemistry term. Suffice to say it’s made of fat. To destroy them we need to break apart that mesh of fat molecules. For this purpose we use the archenemy of fat and grease: soap!
A soap molecule has two parts: a head and a tail. The head is polar and therefore is attracted to water while the tail is non-polar and attracted to oil and fat. When soap molecules are in water they group themselves into micelles – a roughly spherical structure in which all the water-loving heads point outwards (in contact with water) and all the fat-loving tails point inwards at the center of the sphere (away from the water). Therefore it can effectively trap the fat molecule inside the micelle and dissolve the cell membranes.
Precipitating the DNA
When all the membranes are broken, the DNA is released into the solution along with all sorts of proteins molecules and other cellular inclusions. But we cannot take it out yet. Why? It has to do with physical and chemical characteristics of the DNA molecule.
The DNA molecule is a double-helical polymer consisting of a sugar-phosphate backbone with nitrogenous bases running perpendicular to the backbone. These bases often represented by the letters - A (adenine), G (guanine), C (cytosine) and T (thymine) - are the elementary components making up the coded genetic information. The base sequence acts as the instruction manual of the cell, instructing it how to make enzymes and proteins and ultimately everything an organism needs to survive and function.
But we still have not said why the DNA molecules are so soluble in water. It is because of the sugar-phosphate backbone which is highly negatively charged and therefore polar. Since "like dissolves like" water, a polar solvent, will dissolve DNA making it hard for us to take it out of the solution.
To effectively take a polar compound out of a polar solvent, we need to make the compound less polar and reduce its attraction to the solution. Remember that the reason why the DNA is polar is the presence of the negatively charged sugar-phosphate backbone. We need to make it non negative or neutral. To do this, we will add a positively charged ion found in table salt to the solution. Table salt is made of sodium ions (which are positively charged) and chloride ions (which are negatively charged). The sugar-phosphate backbone of the DNA molecule which has a negative charge will be neutralized by the positive sodium ions therefore making the molecule less polar. But it is easier for the negative sugar-phosphate backbone of the DNA to bind with the positive sodium ion when it is in alcohol. That is why we will add alcohol to the solution.
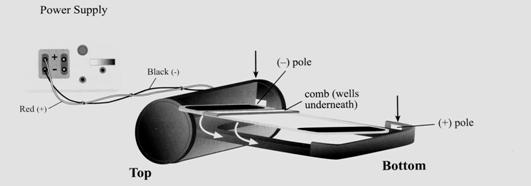
Gel Electrophoresis
After we extract the DNA, we want to run a gel electrophoresis in order to form banding patterns that can help us differentiate two different fruits. Gel electrophoresis is a technique used for separating molecules based on their charge and molecular weight. The sample is loaded in a gel matrix and an electric field is applied across it. Lighter molecules will migrate to the opposite end of the gel faster than heavier molecules. The situation is analogous to swimmers in a swimming pool. The lighter swimmer is able to move faster than the heavier ones and is more likely to reach the end first.
How can molecules move in the gel?
We have learned that DNA is negatively charged due to the phosphate groups in the sugar-phosphate backbone. By applying a voltage difference, creating an electric field across the gel, the negatively charged molecules will move toward the positive side of the field. We assume that the applied electric field is uniform and that all molecules feel the same magnitude of electric force. Think of the uniform electric field as a strong wind blowing at the backs of sprinters pushing each of them evenly.
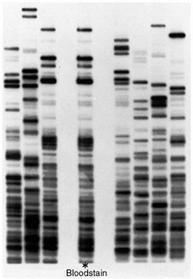
The following is a gel after the samples have been run. Each column is referred to as a lane and they represent one sample each. The individual bands contain fragments of DNA that are identical in weight.
DNA Digestion
One important issue in our case is that the DNA we extract is too large to move through the gel. We have to cut them into smaller fragments. In order to cut DNA molecules into smaller pieces we will digest it using restriction enzymes. These enzymes are highly specific in terms of where in the DNA sequence they cut. There are many different restriction enzymes and each recognizes a different sequence known as a recognition site. For DNA molecules from the same fruit, we will have the same number of fragments of each type.
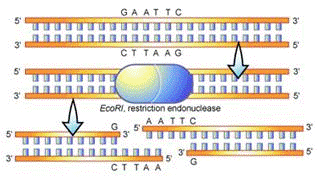
How restriction enzymes work?
We learned earlier that DNA bases are represented by the following letters: A, G, C, and T.
| Enzyme | Source | Recognition Sequence | Cut | |
|---|---|---|---|---|
| EcoRI | Escherichia coli | 5'GAATTC 3'CTTAAG |
5'---G 3'---CTTAA |
AATTC---3' G---5' |
Whenever the enzyme named EcoRI sees the sequence of bases: GAATTC or CTTAAG, it will cut it right between the G and the A.
When a specific enzyme is used a DNA molecule from the same plant will produce the same pattern in the gel. If we use the same enzyme on another plant, we will get another pattern and we can effectively differentiate between the two plants.
3 Your Assignment
Individual Lab Report
Follow the lab report guidelines laid out in the page called Specifications for Writing Your Lab Reports in the Technical Communication section of this manual. As you write, the following discussion points should be addressed in the appropriate section of your lab report:
- Justify the use of salt, soap and alcohol in the extraction procedure.
- Explain how to reach DNA and the barriers to overcome to get to it.
- Describe major techniques used in this lab: Gel Electrophoresis, DNA Digestion with restriction enzymes.
- Explain why the incubator was used.
- Important properties of DNA directly having an impact on the extraction procedure.
- Clearly describe the procedural steps the way they were carried out in lab.
- Describe the steps carried with the TA after you rinse the DNA.
- Describe how is the banding pattern obtained.
- Explain why the banding pattern allow to identify a specific fruit sample.
Team PowerPoint Presentation
Follow the presentation guidelines laid out in the page called EG1004 Lab Presentation Format in the Introduction to Technical Presentations section of this manual. When you are preparing your presentation, consider the following points:
- In your presentation, you need to heavily rely on graphics and pictures.
- Make sure your Experimental Work is described simply and thoroughly.
- Make sure you talk about real-life application of DNA sequencing.
- Demonstrate clear understanding of each procedural step carried out and why it worked.
4 Materials and Equipment
- Fruit Sample
- Non-iodized Table salt (NaCl)
- Hand Soap (clear, unscented)
- 95% isopropyl alcohol (ice cold)
- Distilled water
- Strainer (1 per group)
- Plastic cup (3 per group)
- Ziploc Bag (1 per group)
- Restriction enzyme
- Micropipettes and tips (5 µl and 10 µl)
- Incubator
- Eppendorf tube (1 per group)
- Precast Agarose Gel
- Electrophoresis System
- Blue Digital Bioimaging System
- Foam plate (1 per group)
5 Procedure
- Obtain a fruit sample of about the size of a strawberry. That would be about a quarter of a medium-sized banana.
- Put the fruit sample in the Ziploc bag provided. Carefully, close the bag so there is as little air as possible inside. Start smashing it gently with your hands. Be careful not to burst the bag to avoid a big mess! After about 5 minutes, the fruit sample (banana in most cases) should be transformed into a creamy paste.
- While one team member is working on the homogenization of the fruit sample, another should prepare the buffer solution. Fill the cup half way with distilled water, add about one teaspoon of table salt. Dissolve the salt in the water. Then, add 2 or 3 squirts of soap. Stir it gently so that it doesn’t foam.
- When the desired texture is obtained for the fruit tissue, pour the prepared buffer in the Ziploc bag and close it. Make sure you haven’t trapped air in the bag. Gently mix the smashed fruit and the buffer solution in the bag. Do it slowly. It is important that it does not foam a lot.
- Let the mixture sit for about 2 to 5 minutes. If it has foamed, allow the foam to go away during this time. By letting the mixture stay still, the foam will disappear.
- Use another clear plastic cup to filter the solution. One team member should hold the strainer on top of an empty cup while another carefully pour the content of the Ziploc bag. Make sure it does not foam. Pour slowly! Occasionally shake the strainer to make to liquid filter through. There is a lot of debris.
- Once the solution has been filtered, add ice cold alcohol to it until the total volume reaches ¾ of the cups volume. BECAREFUL, pour the alcohol against the wall of the cup, do not drop all at once. We do not want the alcohol to mix with the solution we want it to float on top. Alcohol is very miscible with water but IT CAN FLOAT IF IT IS POURED SLOWLY AGAINST THE WALL OF THE CONTAINER because it is less dense than water.
- One minute after you have poured the alcohol into the cup, you will see threads of DNA forming. It looks like a translucent gel-like glob at the interface of the juice and the alcohol. Use a paperclip to spool it out of the solution.
- Have 2 other cups 1/4 filled with ice-cold alcohol. Rinse the glob of DNA in the first one for about 10 seconds, then rinse it again the second one for the same time period.
- Allow the rinsed DNA to dry for a minute or two. Call the TA who will guide you with the rest of the procedure: adding the restriction enzymes, putting it in the incubator and running the gel.
- Once you have completed the steps described above, clean your station and follow the TA's instructions. It will take about 30 minutes before you can see the banding pattern formed by your digested DNA in the Gel.
6 References
Vernier Website
Invitrogen Website
Nasco Website
Wikipedia
The Science Creative Quarterly
http://www.hhmi.princeton.edu/documents/labprotocols
http://porpax.bio.miami.edu/~cmallery/255/255chem/255chemistry.htm